Abstract
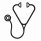
Patients with the plasma cell malignancy multiple myeloma now benefit from treatments such as proteasome inhibitors, immunomodulatory imide drugs (IMiDs), autologous stem cell transplant, and monoclonal antibodies. However, 20% of patients still relapse or die within two years and are deemed 'high risk'. Current markers fail to identify all high-risk patients resulting in misdiagnoses, therefore additional markers for this deadly form of the disease are required.
To better understand and identify high-risk myeloma, we analyzed the structural variant landscape of 826 myelomas from newly-diagnosed patients using whole genome sequencing as part of the CoMMpass trial (NCT01454297). High-confidence somatic structural variants were determined using DELLY and quality control metrics to exclude regions with sequencing anomalies.
Myeloma from newly diagnosed patients had a median of 21 somatic structural variants including 7 duplications, 2 deletions, 7 inversions, and 3 translocations. The number of deletions, duplications, and translocations corresponded to poor progression-free (PFS) and overall survival (OS), with translocations being the most significant (P <6.1x10-7). The two most common translocations occurred at the IgH (41%) and MYC (23%) loci, but did not correspond with differential outcome. However, the third most commonly translocated region (10%) occurred at the IgL locus and was indicative of poor PFS and OS with hazard ratios (HR) of 1.71 and 1.81, respectively (Figure 1). IgL-translocated myeloma did not correspond with distinct clinical features such as age, stage, gender, or b2M levels; and IgL-translocated patients were treated with similar therapeutic regimens as others. Additionally, IgL-translocated myeloma did not have a distinct mutational repertoire, gene expression subtype, and did not have many unique structural genetic elements. The notable exception is that 70% of IgL-translocated myeloma co-occurred with hyperdiploidy, a marker normally associated with better prognosis. However, patients with IgL-translocated and hyperdiploid myeloma experienced poor outcome with a median PFS of 23 months compared to 42.8 months for non-t(IgL) hyperdiploid myeloma (PFS HR=2.35; OS HR=2.41). This poor outcome is partially explained by the failure of patients with IgL-translocated myeloma to benefit from IMiDs. In fact, IMiDs provided no survival benefit to patients with IgL-translocated myeloma who experienced poor outcomes commensurate with patients that did not receive IMiDs (PFS HR=1.56; OS HR=1.49). This is in contrast to patients with myeloma that harbor other translocations, such as IgH translocation, who benefited from IMiDs (PFS HR=0.83, OS HR=0.61).
These data identify IgL translocation as an independent marker of poor prognosis regardless of translocation partner, and suggest this may be due to the failure of this myeloma subtype to benefit from IMiDs. One potential mechanistic explanation is that the IgL enhancer is one of the most robust enhancers of gene expression and is therefore uniquely resistant to therapeutic inhibition. Indeed, the IgL enhancer is bound by several transcription factors at some of the highest levels in the B cell / myeloma epigenome, including BRD4, MED1, and IKZF1. This last factor is particularly interesting as IKZF1 is the target of IMiDs, and thus high-levels of IKZF1 occupancy at the IgL enhancer may be more difficult to fully deplete therapeutically than other loci. This may explain why patients with IgL-translocated myeloma do not benefit from IMiDs whereas patients with IgH- or IgK-translocated myeloma do. Finally, the co-occurrence of myeloma with IgL-translocation and hyperdiploidy is particularly unfortunate, as hyperdiploidy is routinely tested for clinically, whereas IgL-translocations are rarely diagnosed, likely resulting in their misclassification as standard risk.
Figure: IgL translocations portend poor prognosis. a Circos plot showing the repertoire of IgL translocations in newly diagnosed myeloma where line thickness denotes frequency (key bottom left). b Kaplan-Meier analysis of IgL translocated [t(IgL)] patients (N=81) as compared to non-t(IgL) (N=745) for progression-free (PFS; top) and overall survival (OS; bottom). P-values were calculated using a Cox proportional hazards Wald's test or permutation based P-value with 1,000 permutations based on the hazard ratio.
Neri:Celgene: Consultancy, Honoraria; Janssen: Consultancy, Honoraria. Bahlis:Amgen: Consultancy, Honoraria, Research Funding; Celgene: Consultancy, Honoraria, Research Funding; Janssen: Consultancy, Honoraria, Research Funding. Nooka:Adaptive technologies: Consultancy, Membership on an entity's Board of Directors or advisory committees; Amgen: Consultancy, Membership on an entity's Board of Directors or advisory committees; GSK: Consultancy, Membership on an entity's Board of Directors or advisory committees; Takeda: Consultancy, Membership on an entity's Board of Directors or advisory committees; Celgene: Consultancy, Membership on an entity's Board of Directors or advisory committees; BMS: Consultancy, Membership on an entity's Board of Directors or advisory committees; Janssen pharmaceuticals: Consultancy, Membership on an entity's Board of Directors or advisory committees; Spectrum Pharmaceuticals: Consultancy, Membership on an entity's Board of Directors or advisory committees. Kaufman:Janssen: Consultancy; Roche: Consultancy; Karyopharm: Other: data monitoring committee; Abbvie: Consultancy; BMS: Consultancy. Lonial:Amgen: Research Funding. Boise:Abbvie: Consultancy; AstraZeneca: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal